- Research
- Open Access
Respiratory Research 24, Article number: 32 (2023)
Abstract
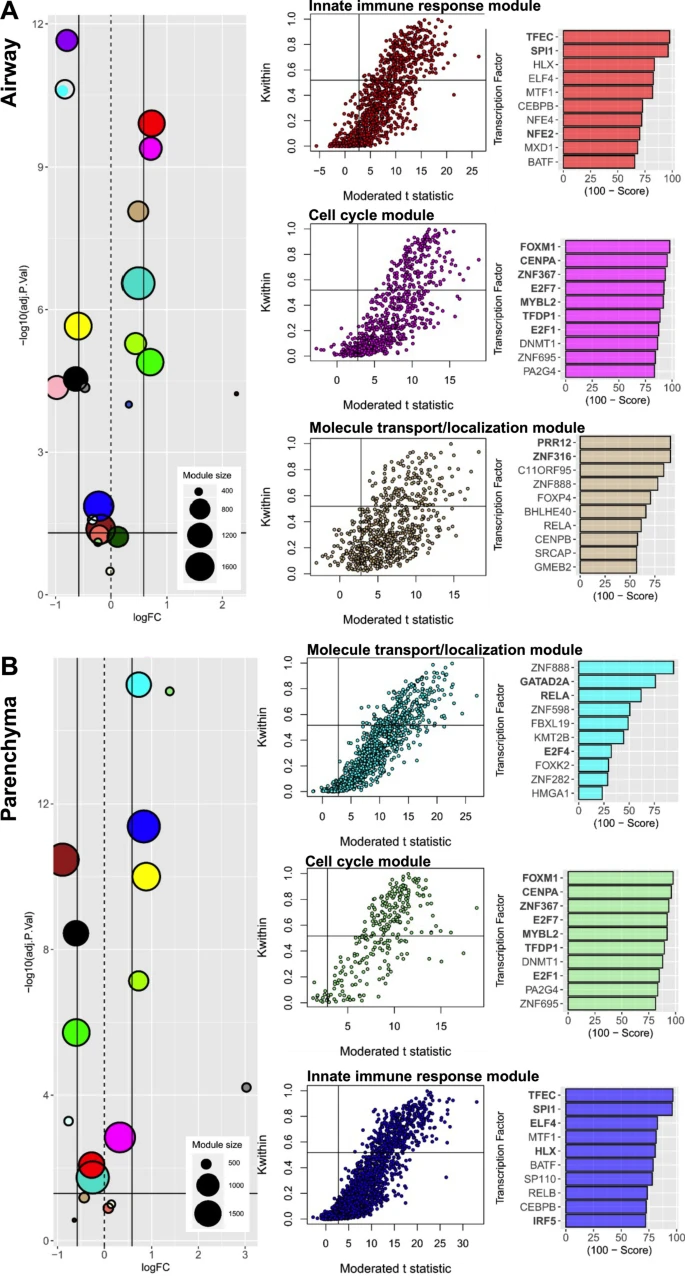
Lung transcriptomics studies in asthma have provided valuable information in the whole lung context, however, deciphering the individual contributions of the airway and parenchyma in disease pathogenesis may expedite the development of novel targeted treatment strategies. In this study, we performed transcriptomics on the airway and parenchyma using a house dust mite (HDM)-induced model of experimental asthma that replicates key features of the human disease. HDM exposure increased the expression of 3,255 genes, of which 212 were uniquely increased in the airways, 856 uniquely increased in the parenchyma, and 2187 commonly increased in both compartments. Further interrogation of these genes using a combination of network and transcription factor enrichment analyses identified several transcription factors that regulate airway and/or parenchymal gene expression, including transcription factor EC (TFEC), transcription factor PU.1 (SPI1), H2.0-like homeobox (HLX), metal response element binding transcription factor-1 (MTF1) and E74-like factor 4 (ets domain transcription factor, ELF4) involved in controlling innate immune responses. We next assessed the effects of inhibiting lung SPI1 responses using commercially available DB1976 and DB2313 on key disease outcomes. We found that both compounds had no protective effects on airway inflammation, however DB2313 (8 mg/kg) decreased mucus secreting cell number, and both DB2313 (1 mg/kg) and DB1976 (2.5 mg/kg and 1 mg/kg) reduced small airway collagen deposition. Significantly, both compounds decreased airway hyperresponsiveness. This study demonstrates that SPI1 is important in HDM-induced experimental asthma and that its pharmacological inhibition reduces HDM-induced airway collagen deposition and hyperresponsiveness.
No comments:
Post a Comment