Qi, C., Li, A., Su, F. et al. Commun Biol 7, 1696 (2024). https://doi.org/10.1038/s42003-024-07416-7
Abstract
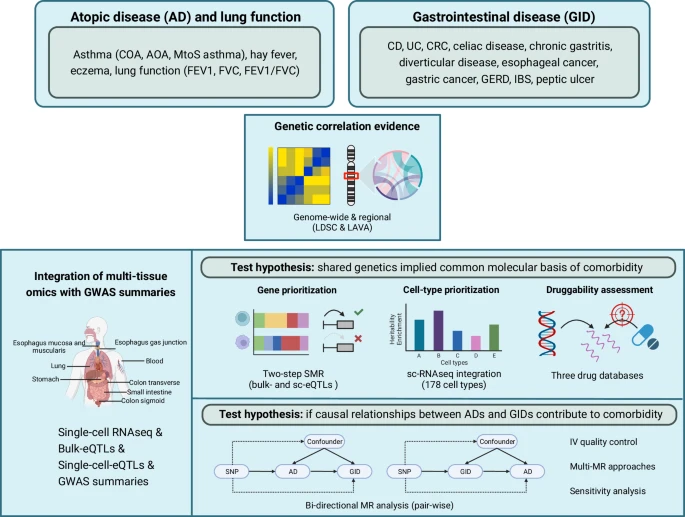 |
| Schematic visualization of the study workflow |
Comorbidity among atopic diseases (ADs) and gastrointestinal diseases (GIDs) has been repeatedly demonstrated by epidemiological studies, whereas the shared genetic liability remains largely unknown. Here we establish an atlas of the shared genetic architecture between 10 ADs or related traits and 11 GIDs, comprehensively investigating the comorbidity-associated genomic regions, cell types, genes and genetically predicted causality. Although distinct genetic correlations between AD-GID are observed, including 14 genome-wide and 28 regional correlations, genetic factors of Crohn’s disease (CD), ulcerative colitis (UC), celiac disease and asthma subtypes are converged on CD4+ T cells consistently across relevant tissues. 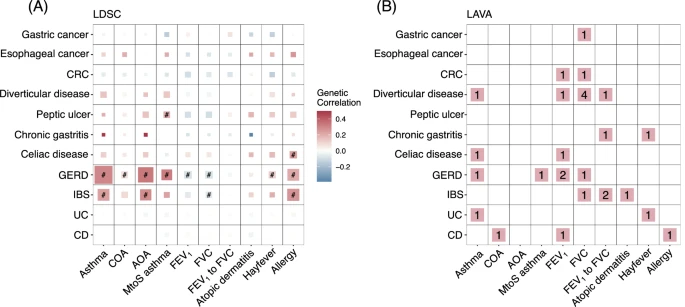 |
| Global and local genetic correlations between AD-GID |
Fourteen genes are associated with comorbidities, with three genes are known treatment targets, showing probabilities for drug repurposing. Lower expressions of WDR18 and GPX4 in PBMC CD4+ T cells predict decreased risk of CD and asthma, which could be novel drug targets. MR unveils certain ADs led to higher risk of GIDs or vice versa. Taken together, here we show distinct genetic correlations between AD-GID pairs, but the correlated genomic loci converge on the dysregulation of CD4+ T cells. Inhibiting WDR18 and GPX4 expressions might be candidate therapeutic strategies for CD and asthma. Estimated causality indicates potential guidance for preventing comorbidity.




No comments:
Post a Comment